Writing processes
Process is a central building block of the Resolwe’s dataflow. Formally, a process is an algorithm that transforms inputs to outputs. For example, a Word Count process would take a text file as input and report the number of words on the output.
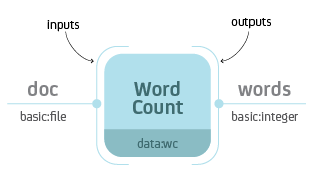
Word Count process with input doc of type basic:file and output
words of type basic:integer.
When you execute the process, Resolwe creates a new Data object with
information about the process instance. In this case the document and the
words would be saved to the same Data object. What if you would like
to execute another analysis on the same document, say count the number of
lines? We could create a similar process Number of Lines that would also take
the file and report the number of lines. However, when we would execute the
process we would have 2 copies of the same document file stored on the
platform. In most cases it makes sense to split the upload (data storage) from
the analysis. For example, we could create 3 processes: Upload Document,
Word Count and Number of Lines.
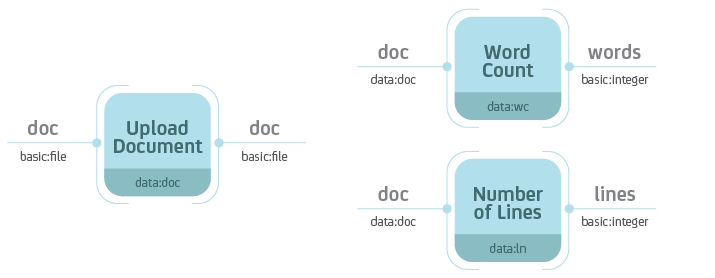
Separate the data storage (Upload Document) and analysis (Word Count,
Number of Lines). Notice that the Word Count and Number of Lines
processes accept Data objects of type data:doc—the type ot the
Upload Document process.
Resolwe handles the execution of the dataflow automatically. If you were to execute all three processes at the same time, Resolwe would delay the execution of Word Count and Number of Lines until the completion of Upload Document. Resolwe resolves dependencies between processes.
A processes is defined by:
Inputs
Outputs
Meta-data
Algorithm
Processes are stored in the data base in the Process model. A process’
algorithm runs automatically when you create a new Data object. The inputs
and the process name are required at Data create, the outputs are saved by
the algorithm, and users can update the meta-data at any time. The
Process syntax chapter explains how to add a process definition to the
Process data base model
Processes can be chained into a dataflow. Each process is assigned a type
(e.g., data:wc). The Data object created by a process is implicitly
assigned a type of that process. When you define a new process, you can specify
which data types are required on the input. In the figure below, the Word
Count process accepts Data objects of type data:doc on the input.
Types are hierarchical with each level of the hierarchy separated by a colon.
For instance, data:doc:text would be a sub-type of data:doc. A process
that accepts Data objects of type data:doc, also accepts Data
objects of type data:doc:text. However, a process that accepts Data
objects of type data:doc:text, does not accept Data objects of type
data:doc.
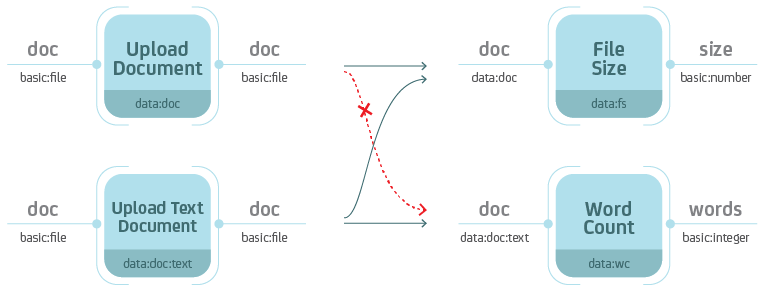
Types are hierarchical. When you define the type on the input, keep in mind that the process should also handle all sub-types.
Process syntax
A process can be written in any syntax as long as you can save it to the
Process model. The most straight-forward would be to write in Python, using
the Django ORM:
p = Process(name='Word Cound',
slug='wc-basic',
type='data:wc:',
inputs = [{
'name': 'document',
'type': 'basic:file:'
}],
outputs = [{
'name': 'words',
'type': 'basic:integer:'
}],
run = {
'bash': 'WORDS=`wc {{ document.file }}\n`' +
'echo {"words": $WORDS}'
})
p.save()
We suggest to write processes in the YAML syntax. Resolwe includes a
register Django command that parses .yml files in the processes
directory and adds the discovered processes to the Process model:
./manage.py register
Do not forget to re-register the process after you make changes to the .yml
file. You have to increase the process version each time you register it. For
development, you can use the --force option (or -f for short):
./manage.py register -f
This is an example of the smallest processor in YAML syntax:
1- slug: mini
2 name: Minimalistic Process
3 requirements:
4 expression-engine: jinja
5 type: "data:mini"
6 run:
7 language: bash
8 program: |
9 echo 'Hello bioinformatician!'
This is the example of the basic Word Count implementation in the YAML
syntax (with the document file as input):
1- name: Word Count
2 slug: wc-basic
3 type: "data:wc"
4 inputs:
5 - name: document
6 type: basic:file
7 outputs:
8 - name: words
9 type: basic:integer
10 run:
11 language: bash
12 program: |
13 WORDS=$(wc {{ document.file }})
14 echo {"words": $WORDS}
If you would like to review the examples of the three processes mentioned above
(Upload Document, Word Count and Number of Lines), follow this
link. Read more about the process
options in Process schema below.
Process schema
Process is defined by a set of fields in the Process model. We will
describe how to write the process schema in YAML syntax. Some fields in the
YAML syntax have different name or values than the actual fields in the
Process model. See an example of a process with all fields. Fields in a process schema:
Field |
Short description |
Required |
Default |
|---|---|---|---|
unique id |
required |
||
human readable name |
required |
||
detailed description |
optional |
<empty string> |
|
version numbering |
optional |
||
data type |
required |
||
menu category |
optional |
<empty string> |
|
automatic grouping |
optional |
||
storage optimization |
optional |
RAW |
|
scheduling class |
optional |
batch |
|
list of input fields |
optional |
<empty list> |
|
list of result fields |
optional |
<empty list> |
|
the algorithm |
required |
||
requirements |
optional |
<empty dict> |
Slug
TODO
Name
TODO
Description
TODO
Version
TODO
Type
TODO
Category
The category is used to arrange processes in a GUI. A category can be any
string of lowercase letters, numbers, - and :. The colon is used to split
categories into sub-categories (e.g., analyses:alignment).
We have predefined three top categories: upload, import and analyses. Processes without this top category will not be displayed in the GenBoard interface, but will be available on the platform.
Entity
With defining the entity field in the process, new data objects will be
automatically attached to a new or existing Entity, depending on it’s parents
and the definition of the field.
entity field has 3 subfields:
typeis required and defines the type of entity that the newDataobject is attached toinputlimits the group of parents’ entities to a single field (dot separated path to the field in the definition of input)descriptor_schemaspecifies the slug of the descriptor schema that is attached to newly created entity. It defaults to the value oftype
Persistence
Use RAW for imports. CACHED or TMP processes should be idempotent.
Scheduling class
The scheduling class specifies how the process should be treated by the scheduler. There are two possible values:
batchis for long running tasks, which require high throughput.interactiveis for short running tasks, which require low latency. Processes in this scheduling class are given a limited amount of time to execute (default: 30 seconds).
The default value for processes is batch.
Input and Output
A list of Resolwe Fields that define the inputs and outputs of a process. A Resolwe Field is defined as a dictionary of the following properties:
Required Resolwe Field properties:
name- unique name of the fieldlabel- human readable nametype- type of field (eitherbasic:<...>ordata:<...>)
Optional Resolwe Field properties (except for group):
description- displayed under titles or as a tooltiprequired- (choices: true, false)disabled- (choices: true, false)hidden- (choices: true, false)default- initial valueplaceholder- placeholder value displayed if nothing is specifiedvalidate_regex- client-side validation with regular expressionchoices- list of choices to select from (label,valuepairs)
Optional Resolwe Field properties for group fields:
description- displayed under titles or as a tooltipdisabled- (choices: true, false)hidden- (choices: true, false)collapsed- (choices: true, false)group- list of process fields
TODO: explain what is field schema. For field schema details see fieldSchema.json.
Run
The algorithm that transforms inputs into outputs. Bash and workflow languages
are currently supported and we envision more language support in the future (e.g.,
directly writing processes in Python or R). Commands should be written to a
program subfield.
TODO: link a few lines from the all_fields.yml process
Requirements
A dictionary defining optional features that should be available in order for the process to run. There are several different types of requirements that may be specified:
expression-enginedefines the name of the engine that should be used to evaluate expressions embedded in therunsection. Currently, only thejinjaexpression engine is supported. By default no expression engine is set, so expressions cannot be used and will be ignored.executordefines executor-specific options. The value should be a dictionary, where each key defines requirements for a specific executor. The following executor requirements are available:docker:imagedefines the name of the Docker container image that the process should run under.
resourcesdefine resources that should be made available to the process. The following resources may be requested:coresdefines the number of CPU cores available to the process. By default, this value is set to1core.memorydefines the amount of memory (in megabytes) that the process may use. By default, this value is set to4096MiB.networkshould be a boolean value, specifying whether the process requires network access. By default this value isfalse.
Types
Types are defined for processes and Resolwe Fields. Data objects have
implicitly defined types, based on the corresponding processor. Types define
the type of objects that are passed as inputs to the process or saved as
outputs of the process. Resolwe uses 2 kinds of types:
basic:data:
Basic: types are defined by Resolwe and represent the data building blocks.
Data: types are defined by processes. In terms of programming languages you
could think of basic: as primitive types (like integer, float or boolean)
and of data: types as classes.
Resolwe matches inputs based on the type. Types are hierarchical, so the same or more specific inputs are matched. For example:
data:genome:fasta:will match thedata:genome:input, butdata:genome:will not match thedata:genome:fasta:input.
Note
Types in a process schema do not have to end with a colon. The last colon can be omitted for readability and is added automatically by Resolwe.
Basic types
Basic types are entered by the user. Resolwe implements the backend handling (storage and retrieval) of basic types and GenBoard supports the HTML5 controls.
The following basic types are supported:
basic:boolean:- booleanbasic:date:- date (format yyyy-mm-dd)basic:datetime:- date and time (format yyyy-mm-dd hh:mm:ss)basic:decimal:- decimal number (e.g., -123.345)basic:integer:- whole number (e.g., -123)basic:string:- short stringbasic:text:- multi-line stringbasic:url:link:- visit linkbasic:url:download:- download linkbasic:url:view:- view link (in a popup or iframe)basic:file:- a file, stored on shared file systembasic:dir:- a directory, stored on shared file systembasic:json:- a JSON object, stored in MongoDB collectionbasic:group:- list of form fields (default if nothing specified)
The values of basic data types are different for each type, for example:
basic:file: data type is a JSON dictionary: {“file”: “file name”}
basic:dir: data type is a JSON dictionary: {“dir”: “directory name”}
basic:string: data type is just a JSON string
Resolwe treats types differently. All but basic:file:,
basic:dir: and basic:json: are treated as meta-data.
basic:file: and basic:dir: objects are saved to the shared
file storage, and basic:json: objects are stored in PostgreSQL
bjson field. Meta-data entries have references to basic:file:,
basic:dir: and basic:json: objects.
Data types
Data types are defined by processes. Each process is itself a data:
sub-type named with the type attribute. A data: sub-type is defined by
a list process outputs. All processes of the same type should have the same
outputs.
Data type name:
data:<type>[:<sub-type>[...]]:
The algorithm
Algorithm is the key component of a process. The algorithm transforms process’s
inputs into outputs. It is written as a sequence of Bash commands in process’s
run.program field.
Note
In this section, we assume that the program is written using the bash
language and having the expression-engine requirement set to jinja.
To write the algorithm in a different language (e.g., Python), just put it in
a file with an appropriate shebang at the top (e.g., #!/usr/bin/env
python2 for Python2 programs) and add it to the tools directory. To run it
simply call the script with appropriate arguments.
For example, to compute a Volcano plot of the baySeq data, use:
volcanoplot.py diffexp_bayseq.tab
Platform utilities
Resolwe provides some convenience utilities for writing processes:
re-importis a convenience utility that copies/downloads a file from the given temporary location, extracts/compresses it and moves it to the given final location. It takes six arguments:
file’s temporary location or URL
file’s final location
file’s input format, which can have one of the following forms:
ending1|ending2: matches files that end withending1orending2or a combination of(ending1|ending2).(gz|bz2|zip|rar|7z|tgz|tar.gz|tar.bz2)ending1|ending2|compression: matches files that end withending1orending2or a combination of(ending1|ending2).(gz|bz2|zip|rar|7z|tgz|tar.gz|tar.bz2)or just with a supported compression format line ending(gz|bz2|zip|rar|7z)
file’s output format (e.g.,
fasta)maximum progress at the end of transfer (a number between 0.0 and 1.0)
file’s output format, which can be one of the following:
compress: to produce a compressed fileextract: to produce an extracted file
If this argument is not given, both, the compressed and the extracted file are produced.
For storing the results to process’s output fields, Resolwe provides a series of utilities. They are described in the Outputs section.
Runtime
TODO: Write about BioLinux and what is available in the Docker runtime.
Inputs
To access values stored in process’s input fields, use Jinja2’s template
language syntax for accessing variables. For example, to access the value
of process’s fastq input field, write {{ fastq }}.
In addition to all process’s input fields, Resolwe provides the following system variables:
proc.case_ids: ids of the corresponding casesproc.data_id: id of the data objectproc.slugs_path: file system path to Resolwe’s slugs
Resolwe also provides some custom built-in filters to access the fields of the referenced data objects:
id: returns the id of the referenced data objecttype: returns the type of the referenced data objectname: returns the value of thestatic.namefield if it exists
For example, to use these filters on the reads field, use
{{ reads|id }}, {{ reads|type }} or {{ reads|name }}, respectively.
You can also use any Jinja2’s built in template tags and filters in your algorithm.
Note
All input variables should be considered unsafe and will be automatically quoted when used in your scripts. For example, the following call:
volcanoplot.py {{ reads.fastq.0.file }}
will actually be transformed into something like (depending on the value):
volcanoplot.py '/path/to/reads with spaces.gz'
If you do not want this behaviour for a certain variable and you are sure
that it is safe to do so, you can use the safe filter as follows:
volcanoplot.py {{ known_good_input | safe }}
Outputs
Processes have three options for storing the results:
as files in data object’s directory (i.e.
{{ proc.data_dir }})as constants in process’s output fields
as entries in the MongoDB data storage
Note
Files are stored on a shared file system that supports fast read and write accesss by the processes. Accessing MongoDB from a process requires more time and is suggested for interactive data retrieval from GenPackages only.
Saving status
There are two special fields that you should use:
proc.rc: the return code of the processproc.progress: the process’s progress
If you set the proc.rc field to a positive value, the process will fail
and its status will be set to ERROR. All processes that depend on this
process will subsequently fail and their status will be set to ERROR as
well.
The proc.progress field can be used to report processing progress
interactively. You can set it to a value between 0 and 1 that represents an
estimate for process’s progress.
To set them, use the re-progress and re-checkrc utilities described
in the Saving constants section.
Resolwe provides some specialized utilities for reporting process status:
re-errortakes one argument and stores it to
proc.errorfield. For example:re-error "Error! Something went wrong."
re-warningtakes one argument and stores it to
proc.warningfield. For example:re-warning "Be careful there might be a problem."
re-infotakes one argument and stores it to
proc.infofield. For example:re-info "Just say hello."
re-progresstakes one argument and stores it to
proc.progressfield. The argument should be a float between 0 and 1 and represents an estimate for process’s progress. For example, to estimate the progress to 42%, use:re-progress 0.42
re-checkrcsaves the return code of the previous command to
proc.rcfield. To use it, just call:re-checkrc
As some programs exit with a non-zero return code, even though they finished successfully, you can pass additional return codes as arguments to the
re-checkrccommand and they will be translated to zero. For example:re-checkrc 2 15
will set
proc.rcto 0 if the return code is 0, 2 or 15, and to the actual return code otherwise.It is also possible to set the
proc.errorfield with this command in case the return code is not zero (or is not given as one of the acceptable return codes). To do that, just pass the error message as the last argument to there-checkrccommand. For example:re-checkrc "Error ocurred." re-checkrc 2 "Return code was not 0 or 2."
Saving constants
To store a value in a process’s output field, use the re-save utility.
The re-save utility requires two arguments, a key (i.e. field’s name) and
a value (i.e. field’s value).
For example, executing:
re-save quality_mean $QUALITY_MEAN
will store the value of the QUALITY_MEAN Bash variable in process’s
quality_mean field.
Note
To use the re-save utility, add re-require common to the
beginning of the algorithm. For more details, see
Platform utilities.
You can pass any JSON object as the second argument to the re-save
utility, e.g.:
re-save foo '{"extra_output": "output.txt"}'
Note
Make sure to put the second argument into quotes (e.g., “” or ‘’) if you
pass a JSON object containing a space to the re-save utility.
Saving files
A convinience function for saving files is:
re-save-file
It takes two arguments and stores the value of the second argument in the
first argument’s file subfield. For example:
re-save-file fastq $NAME.fastq.gz
stores $NAME.fastq.gz to the fastq.file field which has to be of
type basic:file:.
To reference additional files/folders, pass them as extra arguments to the
re-save-file utility. They will be saved to the refs subfield of
type basic:file:. For example:
re-save-file fastq $NAME.fastq.gz fastqc/${NAME}_fastqc
stores fastqc/${NAME}_fastqc to the fastq.refs field in addition to
storing $NAME.fastq.gz to the fastq.file field.
Note
Resolwe will automatically add files’ sizes to the
files’ size subfields.
Warning
After the process has finished, Resolwe will automatically check if all
the referenced files exist. If any file is missing, it will set the data
object’s status to ERROR. Files that are not referenced are
automatically deleted by the platform, so make sure to reference all the
files you want to keep!
Saving JSON blobs in MongoDB
To store a JSON blob to the MongoDB storage, simply create a field of type
data:json: and use the re-save utility to store it. The platform will
automatically detect that you are trying to store to a data:json: field and
it will store the blob to a separate collection.
For example:
re-save etc { JSON blob }
will store the { JSON blob } to the etc field.
Note
Printing a lot ot data to standard output can cause problems when using
the Docker executor due to its current implementation. Therefore, it is
advised to save big JSON blobs to a file and only pass the file name to the
re-save function.
For example:
command_that_generates_large_json > json.txt
re-save etc json.txt
Warning
Do not store large JSON blobs into the data collection directly as this will slow down the retrieval of data objects.